Reference no: EM131469714
1. Download and decompress the sequence data of chromosome 22.
2. Write Perl code to process and analyze the sequence data file downloaded.
a. Read in the data from the file
b. Use regular expression to extract the sequence from the file.
c. Remove non-ATGC characters from the sequence
d. Extract all the open reading frames (ORFs) from the whole sequence.
- An ORF is a part of DNA sequence that has the potential to be translated.
- Its length should be a multiple of 3.
- ORFs are defined as those subsequences which have a start codon 'ATG' and any of the three stop codons 'TAA', 'TAG' and 'TGA'.Each codon includes threenucleotides.
- In addition to start and stop codon, ORFs extracted here should have 30-90 nucleotides.
- Only one stop codon is allowed in each ORF.
e. Print out a message showing how many open reading frames are found in the screen.
f. Translate each ORF into amino acid sequence using the subroutines provided.
g. Write all found ORFsto a new data file.
h. Write all translated amino acid sequence into another new data file.
Importantnotes:
2.a - 2.ishould be done in one .pl file.
Please use the subroutines provided to perform the translation.
Once I implement your function, I would expect to input the data file name from the screen. Shown below is an example.
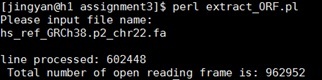
Your output file in step 2.hand 2.i should automatically be created. Below is the example of 2.h output file.
Support information:
1. To write an array into a file, where each entry is shown in one line, use the following command:
print MFILE "$_\n" for @ORFs;
MFILE is the handle for output file.
@ORFs is the array.
2. If you have groups in your pattern
my $string = "TTTATGTGCTGCTAAAAA";
@matches = $string =~ m/^(ATG).*(TAA)$/g
=> With parenthesis () surrounding the subpattern "ATG" ad "TAA", substrings matching the "ATG" and "TAA" part will also be returned.
=> values in @matches will be ("ATGTGCTGCTAA", "ATG", "TAA")
=> Add ?: in the front, such as m/^(?:ATG).*(?:TAA)$/g, substring matching "ATG" and "TAA" will not be returned.
=> In this case the output will be ("ATGTGCTGCTAA")
Download Sequence data of chromosome 22
https://www.dropbox.com/s/6v5whj22boa3kur/hs_ref_GRCh38.p7_chr22.fa.gz?dl=0