Reference no: EM131030777
Genome Assembly
One of the most significant developments of the recent decades was the deciphering of the human genome. The techniques used in this effort can now be used to investigate genetic illnesses, identification of mutations, understanding genomes of ancient organisms that have disappeared, and in numerous other applications.
Genes are encoded within the DNA, a large organic molecule which consists of double chains of four bases: cytosine (cytosine, C), guanine (guanine, G), adenine (adenine, A), and thymine (thymine, T).
Each of the two chains comprises a series of bases, e.g. ACCGTATAG. The other chain consists of bases connected one by one to the first chain bases, with the rules: A-T, C-G. Thus if a chain is ACCGTATAG, the other will be TGGCATATC.
To find the composition of an unknown DNA chain, we work as follows. Using molecular biology techniques, we create multiple copies of the chain and break it into small fragments, for example fragments consisting of three bases each.
Such fragments can easily be identified with special devices. So we end up with a set of known fragments. The problem we then face is to assemble the known fragments into the DNA chain, so that we can learn its exact composition.
Suppose that we have these fragments, or polymersas they are called: GTG, TGG, ATG, GGC, GCG, CGT, GCA, TGC, CAA, AAT, ATG, AAT. Each of them has a length of 3, but generally theycould have a "k" length.To find out from which DNA sequencethey (the fragments) emerged, we create a graph whosevertices are polymers with a length of 2 (or generally k-1),which result from polymers with a length of 3, taking for each polymer with a length of 3, the first 2 (k-1) and the last 2 (k-1) polymers.
Thatway, from GTG wewilltake GT and TG, from TGG we will take TG and GG, etc.
In the graph we add an edge for each one of the initial polymers with a length of 3 (k), from which the current two vertices emerged. We give the name of the initial polymer to the edge. For example, from the polymer ATG we take the vertices AT and ATG and connect them with an edge, which we call ATG. The following illustration shows the graph obtained in this example.
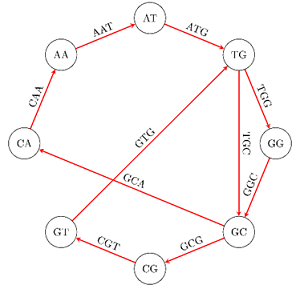
Then, in order to find the original DNA chain, all we have to do is find a path within the graph which visits all the edges exactly once.A path like that is called an Eulerian trail and it uses each edge exactly once.It exists only if at each vertex v, the in-degree of v equals the out-degree of v.
To locate the desired path within the graph we constructed, we use the Hierholzer algorithm:
• We select a random starting node, v.
• We move from node to node until we return to v. The path we have found until now does not necessarily cover all the edges of the graph.
• As long as there is a vertex uthat belongs to the path that we have found but has edges that do not belong to the path:
o We start another path from u, using the edges that have not been used until now, until we get back to u. Then, we connect this path to the path we have already found.
If we use this algorithm on the graph above (picture), we will find the Euler path shown in the figure below:
Then, by traversing the path and joining the nodes, keeping their common base only once, we obtain the DNA sequence ATGGCGTGCA.Note that the sequence is cyclic: the AA node exists in the sequence cycling from the end to the beginning, so we could also take GGCGTGCAAT sequence, or any other that results from rotating the first sequence.
Also, depending on where we start to traverse the path and what choices we make when we have more than one outgoing edges, the starting and finishing points of the sequence can appear to be different.For example, if we start from the TG node, we can obtainthe TGGCAATGCG sequence, or any other that resultsfrom the rotation of this sequence.This does not bother us. The following figures show the cyclical nature of the sequence.
The aim of this assignment is the creation of a program that will assemble DNA chains, using the fragments given.
PROGRAM REQUIREMENTS
Each student will work in his personal repository on GitHub. In order for an assignment to be properly assessed, it must meet the following conditions:
1. The assignment must be contained in a list (folder) called"assignment-2016-2"within the student's repository.
2. The program must be called "dna_assembly.py".
3. The use of ready-made graph libraries or any ready implementations of algorithms or parts of them is strictly forbidden.
4. The program must be called as follows:
python dna_assembly.py fragments_file
The "fragments_file" parameter gives the name of the file in which the fragments are stored (this does not mean that the file MUST be called "fragments_file", the user can give any name he likes).
The file that contains the fragments must have the following format:
ATG
GTG
TGG
GGC
GCG
CGT
GCA
TGC
CAA
AAT
ATG
AAT
That means it has one polymer per line.
Output Description
Caution: if the output is not exactly consistent with its description,the assignment cannot be assessed.
The program should display as an output the DNA sequence that has been identified. So for example, the output will be:
ATGGCGTGCA
or, as mentioned above, another sequence that is equivalent to "ATGGCGTGCA" (since it is cyclic), such as:
TGGCAATGCG
Or
AATGGCGTGC