Genetic Diseases
Problem Description
Many human diseases could be controlled by the knowledge of the gene's structure and pattern. The human gene could be represented by four nucleotides. Each nucleotide is represented by a character - 'J', 'S', 'M', and 'F'. The string pattern of these nucleotides helps us understand the genes. The functions of the genes are based on these strings.
Some biologists want to discover the medicine for the diseases, which could be controlled with the help of these genes. So they decided to compare the two genes and find the problem, so they could make the cure.
Let's have two genes JMFMJFM and MFFJM.
One of the methods to get the problem of these two genes is called alignment comparison. In the alignment comparison, spaces are inserted (if necessary) in some positions of the genes. These spaces make them equally long and then we could score these resulting genes according to the scoring matrix (shown below).
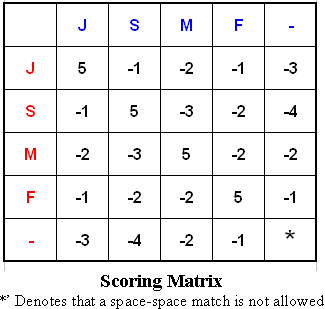
For example, one space is inserted into JMFMJFM to result in JMFMJF-M, and three spaces are inserted into MFFJM to result in -MF--FJM. A space is denoted by the sign (-). The two genes are now equal in length. These two strings are aligned:
JMFMJF-M
-MF--FJM
In this alignment, there are four matches - M in the second position, F in the third, F in the sixth, and M in the eighth. Each pair of aligned characters is assigned a score according to the above scoring matrix.
The score of the alignment above is (-3)+5+5+(-2)+(-3)+5+(-3)+5=9.
Many other alignments are also possible. One is shown below (a different number of spaces is inserted into different positions):
JMFMJFM
-MFFJ-M
This alignment gives a score of (-3)+5+5+(-2)+5+(-1) +5=14.
So, this one is better than the previous one, and this one is also the optimal. No other alignment could give a higher score.
Then, the similarity of the two genes is 14.
This way the max score will result in the least problems.
|
SCORE
|
GRADE
|
|
0 => score
|
0
|
|
0 <= score <= 10
|
A
|
|
10 < score <= 20
|
B
|
|
20 < score <= 30
|
C
|
|
30 < score <= 40
|
D
|
|
40 < score <= 50
|
E
|
On the basis of this grade the biologists could make the cure .
Instructions to work with Open PBT Client:
- Specify the work directory path in the 'Work directory Path' field. The path should correspond to your solution Work directory.
- Download the support files by clicking the Get Dev Files.
- You will find the problem directories containing:
- problem.h file
- problem.c file
in your project directory.
- Code the solution in .c file inside the problem directory
- All required files will be downloaded to your work directory. Creating additional files is strongly discouraged.
Step 1:
In your Solution File:
- Implement your logic in function char rectifyDiseases(char* pattern1 , char* pattern2)
- char* pattern1 : represents the pattern of first gene.
- char* pattern2 : represents the pattern of second gene.
- You can create more functions if required, but those functions should be in the same file.
Step 2:
Your solution needs to consider the following constraints.
- In this problem you have to write a program that helps the biologists to get an accurate grade.
- The human gene could be represented by four nucleotides. Each nucleotide is represented by a character - 'J', 'S', 'M', and 'F'.
- The string pattern of these nucleotides helps us understand the genes.
- The functions of the genes are based on these strings.
- The cure could be prepared only by comparing the two strings.
- One of the methods for comparison is called alignment comparison.
- You have to compare the strings' characters to the Scoring matrix.
- String length should not be greater than 10 else return \0.
- String should be consisting of only given four characters else return \0.
- Characters being returned by the method should be in capital letters.
The Prototype of the function is
char rectifyDiseases(char* pattern1 , char* pattern2) This method takes following arguments.
- pattern1 which represents the pattern of first gene.
- pattern2 which represents the pattern of second gene.
- This method returns a character which is grade.
Constraints
- String length should not be greater than 10 else return \0.
- String should be consist of only given four characters else return \0.
- Characters being returned by the method should be in capital letters.
Example 1
Input
String pattern1=JMFMJFM
String pattern2=MFFJM
Output
B
Example 2
Input
String pattern1=JMFKMJFM
String pattern2=MFFJBM
Output
\0
Explanation: Input string contains other than 'J','S','M' & 'F' characters.
Example 3
Input
String pattern1=JMFMFFFFFFFFFFFJFM
String pattern2=MFFJJJJJJJJJJFFFFFMMMMM
Output
\0
Explanation: Length of the input string is greater than 10.
For C solutions
|
Header File
|
:
|
geneticdiseases.h
|
|
Function Name
|
:
|
char rectifyDiseases(char* pattern1 , char* pattern2);
|
|
File Name
|
:
|
geneticdiseases.c
|
For C++ solutions
|
Header File
|
:
|
geneticdiseases.h
|
|
Class Name
|
:
|
GeneticDiseases
|
|
Function Name
|
:
|
char rectifyDiseases(char* pattern1 , char* pattern2);
|
|
File Name
|
:
|
geneticdiseases.c
|
General Instructions
|
*
|
The file / class names, functions, method signatures, header files are to be used as mentioned in the problem statement. Do not use your own names or change the method signatures and fields. You can add any number of additional methods.
|
|
*
|
For C solutions, change the value of "C_OR_CPP" macro in header file as 1 and for C++ solutions change the value as 2.
|
|
*
|
Incase of iostream.h specify as iostream only.
|
|
*
|
Command line options for the main() function are not supported currently.
|