There are five d-orbitals totally existed. These are designated as dxy, dyz, , dxz, and . The shapes of these orbitals are explained below:
The three orbitals dxy, dyz and dzx are alike and each consists of four lobes of high electron density lying in xy, yz and zx planes correspondingly. These lobes lie in between the principle axes. For instance, in case of dxy, orbital, the four lobes lie in xy plane in between the x- and y-axes.
The 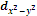 orbital also has four lobes of high electron density along the principle axes x and y.
orbital also has four lobes of high electron density along the principle axes x and y.
The 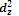 orbital consists of two lobes along the z-axis with a ring of high electron density in the xy plane. The five d-orbitals can be classified into two groups depending upon the nature of their orientation in space.
orbital consists of two lobes along the z-axis with a ring of high electron density in the xy plane. The five d-orbitals can be classified into two groups depending upon the nature of their orientation in space.
The three d-orbitals (dxy, dyz and dzx) which orient in the regions in between the co-ordinate axes are designated as t2g orbitals.
The two other orbitals 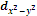 and
and 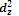 which orient along the axes are labeled as eg orbitals.
which orient along the axes are labeled as eg orbitals.
In the case of a free ion or in an isolated gaseous metal atom/ion all the five d-orbitals have the same energy i.e. they are disintegrate meaning energetically similar. This degeneracy is maintained if a spherically symmetrical field of negative of negative charges surrounds the metal/ion. However, when this negative ends of polar molecules like (H2O, NH3 etc) in a complex, it becomes asymmetrical and the degeneracy of the d-orbitals is lifted. In simple words when the ligand approaches the central metal ion, the electrons in d-orbital of the central metal ion will be repelled by the lone pairs of the ligands. As a result of these interactions the degeneracy of d-orbitals of the metal ions is lost and these split into two set of orbitals having different energies. This is known as Crystal field splitting (Δ) and it forms the basis of crystal field theory.
The energy gap between t2g and eg sets is denoted by Δ0 (octahedral complexes) or Δt (tetrahedral complexes) or 10Dq. The energy difference arises due to difference in electrostatic field exerted by ligands on t2g and eg sets of orbitals of the central metal ion. 10 Dq or Δ is called crystal field splitting energy (CFSE).
It must be noted here that the crystal field splitting largely depends upon the number of ligands crystal field splitting will be different in different structures with different co-ordination numbers.